Analysis and Visualisation Software / BrainVoyager QX
BrainVoyager QX
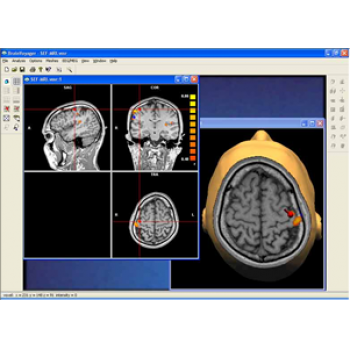
BrainVoyager is the "all-in-one" solution for brain imaging research enables analysis and visualisation of functional and structural magnetic resonance imaging data.
- Highly flexible data processing on raw data
- Comprehensive set of analysis and visualization tools.
- Beautiful visualisations of obtained results.
- Used in more than 500 research groups worldwide, it is known for its high processing speed and interactive, user friendly interface.
- Cross Platform – use your preferred operating system and exploit existing hardware
- Fully open programming interface for advanced users
- Bidirectional connection with BESA (Brain Electrical Source Analysis) allows easy integration with EEG and MEG data.
Guide Price: £4650
SKU: N1600


Supported raw data formats
- Siemens IMA, GE Image, Philips PAR/RED, MR; Bruker, ACR/NEMA, DICOM, ANALYZE
Data preprocessing
- Inter-slice scan time correction (important for event-related fMRI)
- 2D and 3D motion detection and correction
- Spatial and temporal bandpass filtering in frequency space
- Gaussian smoothing in space/time domain
- Removal of linear and higher-order trends in time course data
Statistical analysis
- Parametrical statistical mapping: linear correlation, cross-correlation, General Linear Model (GLM)
- GLM contrasts (t-maps), model comparisons (F-maps)
- Colour-coded relative contribution maps
- Factorial design builder, conjunction analysis, random effects analysis
- Cortex-based fixed – and random-effects GLM
- Volume and Cortex-based Independent Component Analysis (ICA)
- Correction for serial correlations and multiple comparisons, Bonferoni correction, False discovery rate (FDR) correction.
- Region-of interest analysis tool
- Event-related fMRI analysis tools and display
- Granger causality mapping
Utility functions and visualisation tools
- Creation of stimulation protocols; graphical specification of statistical tests; condition-based segmentation and visualization of time course plots
- Simultaneous display of reference function and time course plot
- Protocol-based automatic generation of event-related averaging files
- Display of original data values or percent signal change
- Tabular output for ROI-based GLM analysis
- Loading and display of multiple statistical maps with appropriate map intersection colouring
- Interactive editing of look-up colour tables
- Labeling of anatomical and functional regions
Segmentation and volumetry
- Automatic brain segmentation from 3D data sets using three-dimensional intensity inhomogeneity correction, Talairach templates, histogram analysis, and region growing
- Determination of the volume (mm3) of anatomical regions and functional clusters
- 3D filtering (e.g. Gaussian smoothing) in space or frequency domain
Talairach tools
- Interactive transformation of 3D data sets into Talairach space with visualization of full or partial Talairach grid and coordinates of a selected voxel or the center of gravity of a functional cluster
- Functional 4D time course data in Talairach space allowing: display of time courses of selected voxels or clusters; the computation of statistical 3D maps; inter-subject averaging of 3D anatomical data sets, 4D time courses and 3D statistical maps
Spatial transformation and registration
- Automatic co-registration of functional and anatomical data sets
- Rigid body 3D alignment, 3D motion correction
- Talairach transformation
- Tri-linear and sinc, and cubic spline interpolation
- Interactive multi-modal co-registration (MRI, fMRI, PET, EEG/MEG)
Surface reconstruction
- Head reconstruction with deformable models (i.e. sphere)
- Automatic polygon mesh reconstruction of the segmented cortex along the white/grey matter boundary and the pial surface
- Automatic correction of topological errors in the cortical surface
- Creation and display of multiple surfaces (i.e. scenes composed of cortical hemisphere and transparently rendered head)
- Colouring of convex/concave curvature, sulcal depth and statistical maps
- Automatic cortical volume and cortical thickness analysis
Mesh manipulation
- Interactive real-time slicing through head and brain meshes
- Combined display of multiple cut planes
- Automatic inflation, cutting and flattening of reconstructed cortical hemispheres
- Specification of parameters for various deformation forces
- Reference between 3D coordinates of morphed mesh (e.g. flattened cortex) to original mesh (e.g. folded cortex)
- Display of statistical 3D maps on inflated or flattened surface
- Immediate access and display of time course data for any surface patch
- Surface-based inter-subject alignment
Display utilities
- Simultanous colour-coded display of statistical surface maps or ICA component maps
- Meshes as point models, wire frames, shaded surfaces
- Transparent rendering of selected meshes
- Visualization of MEG/EEG multiple dipole models and waveforms
- Specification of mesh colours and multiple light sources
- Scene antialiasing; scene animation
- Saving and loading of scene viewing conditions
- Creation and display of fMRI and MEG activation movies on slices and meshes
- Export of current 2D or 3D view as 8-bit PNG/GIF/BMP or 24-bit BMP/JPEG file
- Export of dynamic processes as AVI/MNG movie files
- Export of statistical maps merged with 3D structural images as DICOM Exports as .DXF,. VRML or STL polygon meshes (i.e. prototyping, physical brain models)
Coregistration of fMRI with EEG and MEG
- Coregistration of coordinate systems by fiducials and / or surface points
- Direct projection of BESA source models into the individual MRI via interactive link with BV
- Projection of BESA source models into the individual MRI in BESA
- Direct imaging of 3D source images in the individual MRI
- Minimum norm current image based on individual gray/white matter boundary
- Seeding of sources into BESA from anatomical 2D or 3D MR images or from fMRI BOLD clusters in BV via interactive link
- Overlapped display of fMRI and EEG / MEG sources in BV
Hardware requirements
- PC with Intel Pentium II/III/IV, AMD Athlon
- Power Mac G4, G5, Power Book G4, iBook G4
- Unix Workstation (i.e. Sun Solaris)
- Open GL graphics board (i.e. NVIDIA GeForce, ATI Radeon)
- 512 MB RAM
Note: BrainVoyager QX requires a for a single computer or a network dongle providing "floating" licenses. The HASP license system allows you to use the program on Windows, Linux and Mac OS X. With your purchase of BrainVoyager QX, you will receive executables for all these platforms. If you want to use BrainVoyager QX on Unix (i.e. Sun Solaris), please send us an email and we will send you further information about availability of BrainVoyager QX for Unix platforms.